LC/MS/MS Method Package for Primary Metabolites
For LabSolutions™ LCMS Software
LC/MS/MS Method Package for Primary Metabolites Ver.3

Ready-to-use methods for 200 compounds
This Method Package enables efficient, simultaneous analysis of a large number of compounds. Optimized LC separation conditions and MS parameters reduce the time and effort expended on method development.
Choice of optimized analysis conditions
Choose between two method types to suit your analysis aims and equipment. The ion-pair reagent method (112 compounds) is particularly useful for the analysis of sugar phosphates, while the non-ion-pair reagent method (141 compounds), effective in the research of high-value-added substances, does not require ion-pair reagents and in this version has been expanded to include compounds involved in the mevalonate and shikimic acid pathways.
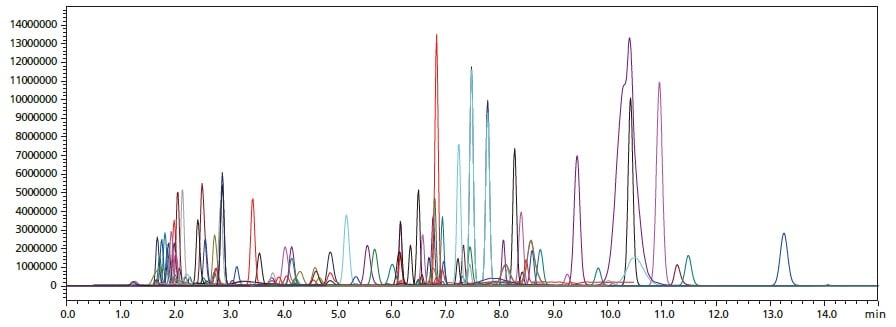
Overlaid MRM chromatograms for simultaneous analysis of a mixture of 141 standards with the non-ion-pair reagent method
Visualization of metabolic changes
With the included Multi-omics Data Analysis Package, quantitative data can be visualized easily on a metabolic map.
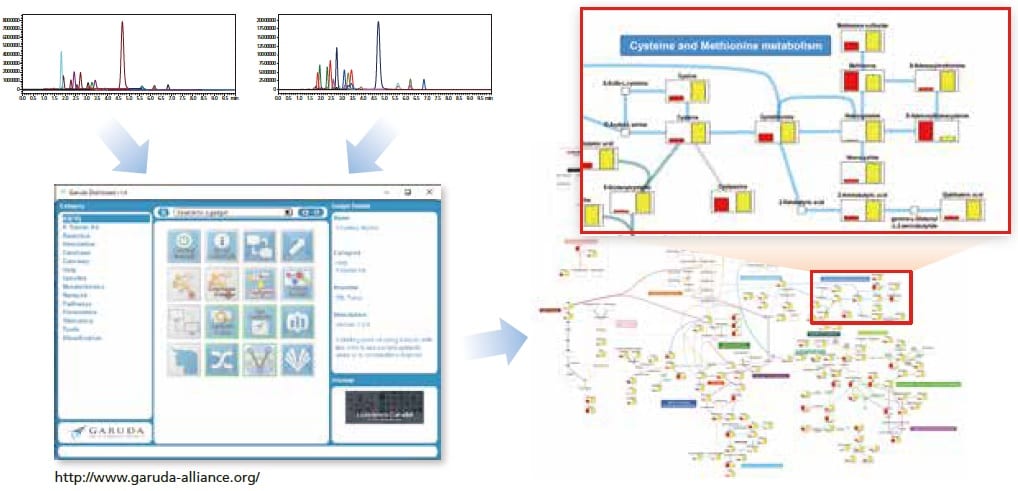
Visualization of simultaneous analysis results from the PFPP column method, created with the Multi-omics Data Analysis Package
Compatible with the Nexera™ series and the LCMS-8060NX
All methods are compatible with both the Nexera series and the LCMS-8045/8050/8060 (NX), covering a wide range of analysis needs.
A total solution from pretreatment to analysis
This Method Package includes protocols for preparing biological tissue extracts, enabling stable analysis with proven pretreatment techniques and reducing the labor and expense involved in method development.
Index of compounds
| List of compounds for ion-pair reagent method | ||
| Glycolytic system | Co-enzyme | Nucleotides |
| 2,3-Bisphosphoglyceric acid 3-Phosphoglyceric acid (2-Phosphoglyceric acid) Dihydroxyacetone phosphate Fructose 1,6-bisphosphate Glucose 1-phosphate Glucose 6-phosphate Glycerol 3-phosphate Phosphoenolpyruvic acid Pyruvic acid |
3-Hydroxybutyryl coenzyme A Butyryl coenzyme A Coenzyme A Crotonyl coenzyme A FAD FMN Malonyl coenzyme A Methylmalonyl coenzyme A NAD NADH NADP NADPH Nicotinic acid Pyrroloquinoline quinone |
Adenine Adenosine Adenosine 3',5'-cyclic monophosphate Adenosine diphosphate Adenosine monophosphate Adenosine triphosphate AICAR Cytidine Cytidine diphosphate Cytidine monophosphate Cytidine triphosphate Guanine Guanosine Guanosine 3',5'-cyclic monophosphate Guanosine diphosphate Guanosine monophosphate Guanosine triphosphate Inosine Inosine monophosphate Orotic acid Thymidine Thymidine diphosphate Thymidine monophosphate Thymidine triphosphate Thymine Uridine Uridine diphosphate Uridine monophosphate Uridine triphosphate Xanthosine monophosphate |
| Pentose-phosphate pathway | ||
| Fructose 6-phosphate Glyceraldehyde 3-phosphate 6-Phosphogluconic acid Erythrose 4-phosphate Ribose 1-phosphate Ribose 5-phosphate Ribulose 5-phosphate Sedoheptulose 7-phosphate |
||
| Sugar phosphate | Nonmevalonic acid pathway | |
| Fructose 1-phosphate Mannose 6-phosphate Phosphoribosyl pyrophosphate Ribulose 1,5-bisphosphate |
HMBPP IPP_DMAPP MEP |
|
| TCA cycle | Shikimic acid pathway | |
| Acetyl coenzyme A 2-Ketoglutaric acid Succinyl coenzyme A |
Shikimic acid Shikimic acid 3-phosphate |
|
| Amino acids | Organic acids | Nucleotide sugar |
| 2-Aminobutyric acid 4-Aminobutyric acid Alanine Arginine Asparagine Aspartic acid Cysteine Glutamic acid Glutamine Glycine Histidine 4-Hydroxyproline Isoleucine Leucine Lysine Methionine Phenylalanine Proline Serine Threonine Tryptophan Tyrosine Valine |
2-Isopropylmalic acid 3-Hydroxyphenylacetic acid 4-Hydroxyphenyl pyruvic acid Citramalic acid Glyceric acid Glycerol 2-phosphate Glycerol 3-phosphate Glycolic acid Indole 3-acetic acid Pantothenic acid |
ADP-glucose UDP-glucose |
| Purine derivative | ||
| Hypoxanthine Uric acid Xanthine |
||
| Internal STDs | ||
| 2-Morpholinoethanesulfonic acid Methionine sulfone |
||
| List of compounds for PFPP column method | ||
| Glycolytic system | Organic acids | Co-enzyme |
| Lactic acid Pyruvic acid |
2-Aminobutyric acid 4-Aminobenzoic acid 4-Aminobutyric acid Caffeic acid Cholic acid Creatine Ferulic acid Glycolic acid Glyoxylic acid Ophthalmic acid Orotic acid p-Coumaric acid Phenyllactic acid Phenylpyruvic acid Taurocholic acid Urocanic acid Vanillic acid |
FAD FMN NAD Niacinamide Nicotinic acid |
| TCA cycle | Nonmevalonic acid pathway | |
| 2-Ketoglutaric acid Aconitic acid Citric acid Fumaric acid Isocitric acid Malic acid Succinic acid |
DOXP MEP |
|
| Urea cycle | Catecholamine | Mevalonic acid pathway |
| Argininosuccinic acid Ornithine Citrulline |
Dopamine Epinephrine Norepinephrine Serotonin |
Mevalonic acid MVA-P |
| Amino acids | Vitamin B | Alkaloid |
| 4-Hydroxyproline Alanine Anthranilic acid Arginine Asparagine Aspartic acid Asymmetric dimethylarginine Cystine Dimethylglycine Glutamic acid Glutamine Glycine Histidine Homocystine Isoleucine Leucine Lysine Methionine sulfoxide Phenylalanine Proline Serine Symmetric dimethylarginine Threonine Tryptophan Valine |
Folic acid Pantothenic acid PLP |
Higenamine Reticuline THP |
| Nucleosides and Nucleotides | Others | |
| Adenine Adenosine Adenosine 3',5'-cyclic monophosphate Adenosine monophosphate Adenylsuccinic acid AICAR Cytidine Cytidine 3',5'-cyclic monophosphate Cytidine monophosphate Cytosine Guanine Guanosine Guanosine 3',5'-cyclic monophosphate Guanosine monophosphate Inosine Thymidine Thymidine monophosphate Thymine Tyrosine Uracil Uridine |
4-Aminophenylalanine 4-Aminophenylpyruvic acid 4-Hydroxybenzoic acid Acetylcarnitine Acetylcholine Carnitine Carnosine Catechol Choline Citicoline Creatinine Cysteamine Dihydroxyphenylacetaldehyde Dihydroxyphenylacetic acid Dopa Ergothioneine Histamine Histidinol Hydroxytyrosol Indole Kynurenine Methyl-DOPA Protocatechuic acid Protocatechuic aldehyde Resveratrol Salicylic acid Sinapic acid Tyramine Vanillin |
|
| Methylation and Transsulfuration cycle | Purinederivative | Internal STDs |
| Cystathionine Cysteine Homocysteine Methionine 5-Glutamylcysteine Glutathione Oxidized glutathione S-Adenosylhomocysteine S-Adenosylmethionine |
Allantoin Hypoxanthine Uric acid Xanthine |
2-Morpholinoethanesulfonic acid Methionine sulfone |
| Shikimic acid pathway | ||
| 3-Dehydroquinic acid 3-Dehydroshikimic acid Chorismic acid Shikimic acid Shikimic acid 3-phosphate |
||
* With this method package, choose between the ion-pair reagent method (112 compounds) or the PFPP column method (141 compounds) depending on your equipment and analysis aims.
Precautions
- LabSolutions LCMS Ver.5.99 SP2 or later is required.
- This method package is for research use only..
LabSolutions and Nexera are trademarks of Shimadzu Corporation.
GARUDA is a trademark of The Systems Biology Institute.
Applications
| Documents | Date Creation Date |
|---|---|
2019-11-19 | |
2019-10-23 | |
2019-08-28 |
News / Events
-
New High Resolution Accurate Mass Library for Forensic Toxicology
Perform forensic toxicology screening for drugs of abuse, psychotropic drugs, pharmaceuticals, pesticides, and natural toxins using this high-resolution accurate mass database.
-
Shimadzu has released the Shim-vial™ H glass, S glass.
Shimadzu provides high-quality vials that thoroughly eliminate these risks by visually inspecting each vial, allowing them to be used with confidence.
-
INTERNATIONAL MASS SPECTROMETRY CONFERENCE 2024
Visit the Shimadzu booth at the International Mass Spectrometry Conference (IMSC) 2024.
-
Metabolomics 2024
Shimadzu Lunch Presentation at Metabolomics 2024
Date: junio 20th, 2024 (Thursday)
Time: 12:25 – 1:25 p.m. -
New LabSolutions Insight Biologics
Supporting Innovation with Oligonucleotide Therapeutics
-
Shimadzu has released the LCMS-TQ RX Series High-Performance Liquid Chromatograph Mass Spectrometers
Innovative technology, exceptional design and new ways of thinking are part of our engineering DNA delivering solutions for the ever-changing needs of any laboratory. As our scientific and business needs change our engineering design evolves and adapts. The result is the RX Series of triple quadrupole LC-MS instruments designed with unmatched capability, redefined reliability and creating a new standard in actionable data.





